For instructions on how to use this pipeline for FRB detection, refer to the Observation Instructions. Remember to update frb.cat for any new FRBs, and to update the DM, RA, DEC as improved numbers come out.
Observation logs are stored in /data/frb/obs_logs{source_name}. Retain the .fil files for a few days in case another telescope detects an FRB. Before deleting a day's worth of data, ensure to check /data/frb/{date}/good for any unreported detections.
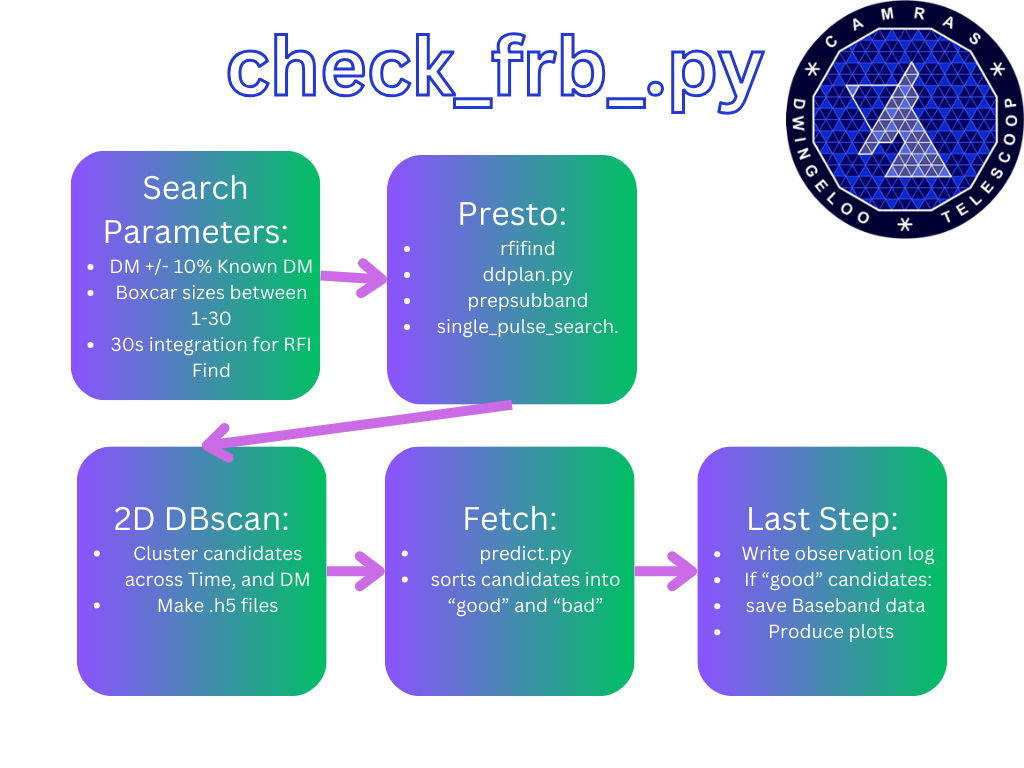
The pipeline records data across four bands: PH, PV, L1, and L2, saving them as .fil files. Once a .fil file is complete, check_frb.py initiates a search over the file, filtering out RFI and searching for single pulses in a dedispersed time series using presto. After the single pulse search, candidate clustering reduces duplicates. fetch then sorts the candidates into "good" or "bad." If "good" candidates are found, baseband data for all bands are saved. We record data in chunks of 10 minutes.
- SNR Detection Threshold: 6
- Integration Time: 30s for
rfifind - Boxcar Sizes: [1, 2, 3, 4, 6, 9, 14, 20, 30] for
single_pulse_search.py - DBscan Clustering: ddm=0.0001, eps=0.01, min_samples=3
- Candidate Threshold: If clustering results in more than 800 candidates, the file is skipped.
- Fetch Model: "m", with a 50% probability threshold
- Baseband Data: 2 seconds recorded for each "good" detection
- Small File Deletion:
.filfiles under 50 MB are deleted without being searched.
This startup script allocates 12 CPU cores per instance of check_frb. It performs the following:
- Runs
/get_frb_from_pointing.pyto verify the telescope is on target. - Checks if data recording is in progress; if so, it takes no further action.
- Initiates filterbank data recording in
screensessions named "filterbank_*band". - Starts
clear_ram_dir, which deletes files older than 15 minutes from/data_tmp. - Initiates
usrp-lband.shand SSH's intocamrasdemo@mercuriusto startusrp-pband.shinscreensessions. - Launches
frb_dashboard.py.
This primary script searches .fil data for FRBs, saves baseband data, and generates diagnostic plots.
Refer to the Presto Tutorial. Presto calls include:
-
DDplan.py: Creates a dedispersion plan.DDplan.py -d {high_dm} -l {low_dm} -n {numchan} -b {BW} -t {dt} -f {fctr}- High and low DM are +/- 10% of the target DM in
frb.cat.
-
rfifind: Filters out RFI.rfifind -ncpus {ncpu} -o {outname} {filterbankfile} -time {time}- 30s default for RFI find.
-
prepsubband: Dedisperses the data."prepsubband -ncpus {ncpu} {noclip_option} {zerodmoption} {ignorechanoption} {rfifindoption} -nsub {nchan} -lodm {low_dm} -dmstep {str(dDM)} -numdms {n_DMS} -o {outname} -nobary {filterbankfile}"
-
single_pulse_search.py: Searches the dedispersed data."single_pulse_search.py -b -t {threshold} {outname}*.dat"- Outputs to "{basename}_DM*.singlepulse".
dbscan_clustering.py clusters candidates across time and DM space. If more than 800 candidates are found after clustering, the file is ignored. If less, .h5 candidate files are generated, and a log file is created.
fetch uses machine learning to classify candidates as "good" or "bad". It ensures only one instance runs at a time using a lockfile, moves the files accordingly, and saves baseband data along with diagnostic plots. Located in the run_predict_and_move function using a lock file.
Located in the run_predict_and_move function, baseband data is saved if "good" candidates are found by fetch. It saves 2 seconds of data centered on the candidate and attempts to save corresponding data in other bands. Saved in .sigmf format.
After saving baseband data, diagnostic .png files are generated for "good" .h5 candidates. Baseband data, good candidate files, and diagnostic plots are located in /data/frb/{date}/good
Logs are written after DBscan clustering to /data/frb/obs_log/{source_name}. The logs include details about the number of candidates before and after clustering, along with other relevant information.
- `#Filterbank_File", "User", "Timestamp", "RFI_Instances", "Pulse_Candidates", "Clusters_(Candidates_after_clustering)", "silhouette_score", "duration_seconds"
- `.fil` file headers are recorded here
This script ends the program, terminating all top-level screen sessions initiated by start_frb.sh. It does not kill screen sessions running check_frb or writing baseband data.
- Automate posting "good" candidates from fetch to the Camras Mastodon bot.
- Update the
frb.catfile to.keyformat. - Try out interferometry using baseband data of the Crab. (Before a real FRB).
- Improve DBscan clustering by considering detection brightness. (IE 3D)
- Experiment with
TransientxoverPRESTOfor faster pipeline processing. - Make a desktop application for easier program initiation on Mercurius.
- Implement automatic shutdown when storage on
/data/frbis below 20GB. Run this in a screen session. - Add a poster outside the telescope.
- Test pipeline with simulated FRBs injectioned into
.filfiles. - Investigate potential misclassification of bright FRBs as RFI. See this plot where there are missing high SNR detections.
- Baseband data and their associated
screensessions are not terminated when the program ends. - Observing past midnight does not automatically create a new
/data/frb/{date}directory. .filfile names use local time instead of UCT, although headers are correct.- Recording of
.filfor L-Bands occasionally crashes - Currently the pipeline is limited by the abililty to write
.filand baseband files
To use this pipeline, install Fetch and Presto. Note: Our version of presto uses a modified ddplan.py to suppress plotting.
Contributions are welcome! Contact @dijkema to get involved.
We welcome feedback on bugs, feature suggestions, and more. Reach out to us for any comments.
Thanks to Astron & Jive for hiring Max Fine under the guidance of Dr. Dr. Tammo Jan Dijkema and Professor Jason Hessels for this summer 2024 project.