This project focuses on analyzing three-dimensional morphology of blood vessels in neoplastic tumors using the 3D Power Doppler ultrasound technique. This imaging method enables detailed visualization of tumor vascularization and facilitates monitoring the effects of anticancer therapies on vessel morphology and quantity.
The primary objective of this project is to develop an algorithm capable of analyzing morphological features of vessels based on Doppler ultrasound images.
The algorithm comprises several processing steps:
- Binarization – Conversion of grayscale images into binary representations.
- Thresholding – Selection of vessel structures based on intensity values.
- Skeletonization – Reduction of vessels to their central lines for morphological analysis.
- Tree Building – Construction of vessels structure representation. 🌳
- Post-Processing – Refinement of the vascular model by eliminating noise and artifacts. 🔧
- Feature Extraction – Calculation of morphological parameters. 🔍
The algorithm provides information on the following features:
- Vessel-Volume Ratio: Proportion of vessel volume to total tumor volume.
- Number of Vascular Trees: Count of distinct vascular structures. 🌳
- Vessel Length: Total length of all vessels in the image.
- Number of Branches: Total branch points in the vascular network. 🌿
- Number of Cycles: Count of closed loops in the vascular structure. 🔄
- Cycle Size: Size of closed loops in the vascular structure. 🔁
- Tortuosity Measurements: Metrics to quantify vessel curvature and irregularity. 🌀
The developed algorithm has been applied to study the normalization process of neoplastic vessels influenced by metformin therapy and oxygen microbubbles. These tools provide insights into how treatments impact vascularization, aiding the development of more effective cancer therapies.
The program supports the analysis of both DICOM and TIFF image formats. Additionally, it can process metadata associated with these images from accompanying XML or CSV files. 📁
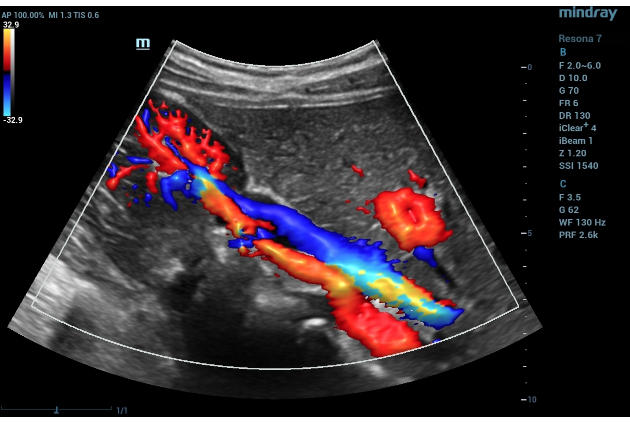
Below is an example of an input image:
The tool features a dedicated Tkinter-based GUI that simplifies the analysis process, making it more user-friendly and accessible. 🎨
Screenshot of the user interface:

Here is a research article that uses this program
Link to Research Article